The DSI Research Software Development Office supports researchers to build new tools or to refine existing software tools to improve usability and robustness and enhance existing functionality.
Only open-source projects are considered for support so that we can disseminate research software beyond the research space in which it is created.
For further information and timelines see Research Software Development Program.
A web application that helps international and domestic university students to develop academic language competencies and foster cross-cultural knowledge exchange through a digital platform.
Researcher: Eunice Jang, U of T, OISE
DSI Software development office: Prototyping, code and architecture review.
Profs. Laura Rosella (Dalla Lana School of Public Health, University of Toronto) and Birsen Donmez (Department of Mechanical & Industrial Engineering, University of Toronto) developed an RShiny app that enables non-coding users to upload data and view analysis results via a web portal. The app’s focus is on the Chronic Disease Population Risk Tool (CDPoRT), which utilizes population-level health system data to predict future chronic disease burden based on individual characteristics. The tool provides data-driven support, guiding resource allocation and improving public health planning.
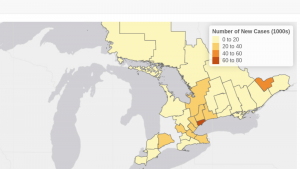
To enhance accessibility and usability, the investigators sought research software support to create a new web interface for the existing algorithm. The DSI Research Software Development played a crucial role in designing the new web interface, developing new code, creating associated documentation, and performing prototyping. Their collaborative efforts ensure that the tool is easily accessible online, allowing researchers and decision-makers to benefit from the data-driven insights provided by the CDPoRT.
GitHub can be viewed here
An interactive map of Toronto cycling infrastructure, with options to view optimal designs, add routes, and calculate accessibility scores.
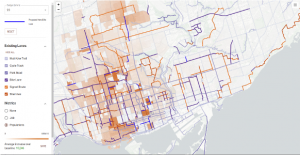
Researcher: Shoshanna Sax, Faculty of Applied Science and Engineering; Timothy Chan, Faculty of Applied Science and Engineering
DSI Software development office: Design, coding, deployment.
A new web portal that will allow physicians to log in and view the reports assigned to them. There is also a backend and admin portal from which the Gemini team will be assigning reports and managing the portal
Researcher: Amol Verma, Unity Health Toronto
DSI Software development office: Implementation end to end including deployment.
Study of men’s and women’s positioning in global scientific collaboration networks. Data analysis project using Elsevier’s Scopus database. Topic is gender within context of research collaboration networks.

Researcher: Megan Frederickson, U of T
DSI Software development office: Consulting, data transformation.
Digital humanities. Goal is a searchable database of TEI-coded medieval manuscripts.
Researcher: Dorothea Kullman, U of T
DSI Software development office: Consulting (GitHub, TEI, project structure).
Prof. Benjamin Haibe-Kains (University Health Network) looked to build upon ORCESTRA, a cloud-based platform to provide an automated and streamlined data processing pipeline submission process to accelerate the pipeline integration process, thus greatly expanding the catalog of the curated data Biomedical objects. This data is used in cancer research have been used to, among many, to identify biomarkers of drug and radiation response, as well as potential toxicities.

The DSI Research Software Development team consulted on the software architecture and recommend potential enhancements so that the software can be extended to a multitude of data types. The team is also working on enhancing data security, conducting a thorough review of the existing architecture within the data processing layer to identify areas for improvement, and considering the reengineering of the pipeline submission feature with a potential migration to the cloud environment.
GitHub can be viewed here
Dr. Angeliki Veroniki (Unity Health Toronto) aimed to develop a user-friendly web interface called Rank-Heat Plot R Shiny tool. This tool allows health researchers to upload spreadsheets containing medical treatment results and visualize and compare outcomes easily. It particularly addresses the challenges faced by health researchers, especially in data interpretation, when utilizing network meta-analysis (NMA) that combines evidence from multiple randomized trials to compare various treatments.
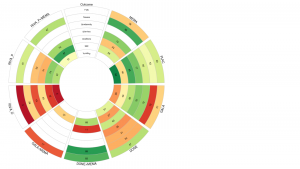
The DSI Research Software development team created the R Shiny interface for the Rank-Heat Plot tool. They updated existing R code as necessary and collaborated with lab members to deploy the application on an on-premise servers or in the cloud, depending on requirements. The tool can now empower clinicians, guideline developers, and policy makers to make well-informed decisions regarding drug coverage, provide recommendation and discuss optimal treatment options with patients across different outcomes.
An open-source dynamics package which is a collection of numerical integrators and tools to enable dynamical simulations of bodies such as stars, planets, asteroids, and comets.
Researcher: Hanno Rein, University of Toronto Scarborough
Github: https://github.com/hannorein/rebound
DSI Software development office: Bug fixes and enhancements. Containerizing the application and implementing the visualization widget
Prof. Bo Wang (Laboratory Medicine & Pathobiology, Temerty Faculty of Medicine, University of Toronto) developed scGPT, a GPU-powered tool that empowers non-technical users to conduct RNA-seq analysis. It leverages a pre-trained machine learning model and enables tasks like gene expression profiling, differential gene expression analysis, clustering, and more.

The DSI Research Software Development team were instrumental in the development and deployment of scGPT by contributing to the coding of the application, ensuring its functionality and usability. They facilitated the deployment of the tool, making it accessible to users through a web-based interface.
Temporary development site can be viewed here
Developing a new database app for sample images, sample metadata, and analysis results.
Researcher: Monica Ramsey, UTM
DSI Software development office: Code, documentation, deployment.
Link to project: www.ramseylab.ca
Software that explicitly accounts for segmentation errors when attempting to cluster single-cell data from highly multiplexed imaging.
Researcher: Kieran Campbell
DSI Software development office: coding, documentation, testing, package development
Prof. Ewan Dunbar (Department of French, Faculty of Arts & Science, University of Toronto) recently introduced a prototype web interface called Speech Features Online. This interface serves as a platform for speech researchers to conveniently upload their audio files and access downloadable speech features essential for speech processing tasks. The software holds significant value for both experimental and clinical speech researchers, aiding them in their respective endeavors.
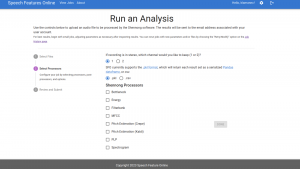
To bring this prototype to life, the DSI Research Software Development team took charge of developing the required code and documentation for implementing the new tool. Their expertise and efforts were instrumental in transforming the concept into a functional and user-friendly web interface.
Widget-enabled Jupyter notebook enabling non-technical users to run existing algorithms easily.
Researcher: Leo Chou, U of T
DSI Software development office: Code, deployment.
Dr. Gregory Schwarz (University Health Network) had a vision to deploy a set of tools for clustering and visualizing single-cell data. To bring this vision to life, the DSI Research Software Development team developed an app as a reimplemented version of an existing static visualization algorithm. The team was responsible for code development and documentation to ensure the tool’s functionality and usability.
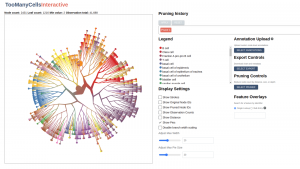
One notable component of TooManyCells is its custom-made visualization feature. The DSI Research Software Development team designed this component, which presents cell relationships as a tree. Through the utilization of TooManyCells, it has become possible to identify rare cancer cells that contribute to disease progression, thus addressing an important need in the field. As a result of their efforts, an article describing the tool has been submitted for publication.
GitHub can be viewed here
Featured in DSI news here
Link to publication here
Zoobot3D is a cutting-edge new DSI-funded software development project that connects AI industry with human ingenuity, efficiently measuring, labelling, annotating and cataloguing images of deep space. Zoobot3D will be the first and only software tool for galaxy feature segmentation, underpinning a new field of research that will help researchers answer questions that would otherwise be impossible.
Researcher: Jo Bovy (David A. Dunlap Department of Astronomy and Astrophysics, Faculty of Arts & Science, University of Toronto); Joshua Speagle (Department of Statistical Sciences, Faculty of Arts & Science, University of Toronto)
Github link here.
Featured in DSI News here.
The University of Toronto provides resources for data infrastructure. You can view a complete set of resources at the Centre for Research Innovation & Support Digital Research Infrastructure Portal
A Data Management Plan (DMP) contains information on how the research team intends to handle research data both during the research project and after the project is complete. Many research sponsors require a DMP as part of the application process.
SciNet is the supercomputer centre at the University of Toronto, hosting Niagara, one of the fastest supercomputers in Canada, plus a range of other systems. SciNet provides Canadian researchers with computational resources and expertise necessary to perform their research on scales not previously possible in Canada.
SciNet is a suitable storage and compute location for UK BioBank data, and researchers can list SciNet in the data management plan of their UK Biobank and Research Ethics Board applications.
There are a variety of data storage options available locally at the University of Toronto and externally. Researchers can select the storage option based on their specific requirements.
Use this tool to find further options.