Tumors, much like people, are different from one another. In fact, not only can the same type of tumor vary from person to person, but there can also be variations within the tumor itself, as a single tumor is comprised of a diverse population of cells. This tumor heterogeneity makes it difficult for researchers to create effective treatment plans. This is where Dr. Gregory Schwartz and his team at the University Health Network, and Medical Biophysics at the University of Toronto, come in with the help of the Data Sciences Institute’s (DSI) research software development support program.
Interested in applying for the DSI’s research software development support program? Apply by October 21, 2022, for our next round of applications.
The DSI’s software development program is designed to support faculty and scientists by providing access to highly skilled software developers to refine or enhance existing software and improve usability and robustness, build new tools, and disseminate research software. The DSI has been supporting six projects since its first call. The DSI’s senior software developer, Dr. Conor Klamann worked with Schwartz and his team.
Helping understand cellular heterogeneity in cancer with TooManyCells
Genetic heterogeneity within a tumor occurs due to imperfect DNA replication. When healthy cells divide to create new cells, it can lead to mutations. When cancerous cells divide, mutations can also occur causing tumor heterogeneity. However, these diverse populations of cells can also exhibit non-genetic heterogeneity in response to treatment, changing their behaviour based on their surrounding environment independent of mutation. To measure cell behavior at the resolution of individual cells, researchers are using new single-cell technologies. This produces a massive amount of detailed data and subsequently requires sophisticated computational tools to interpret.
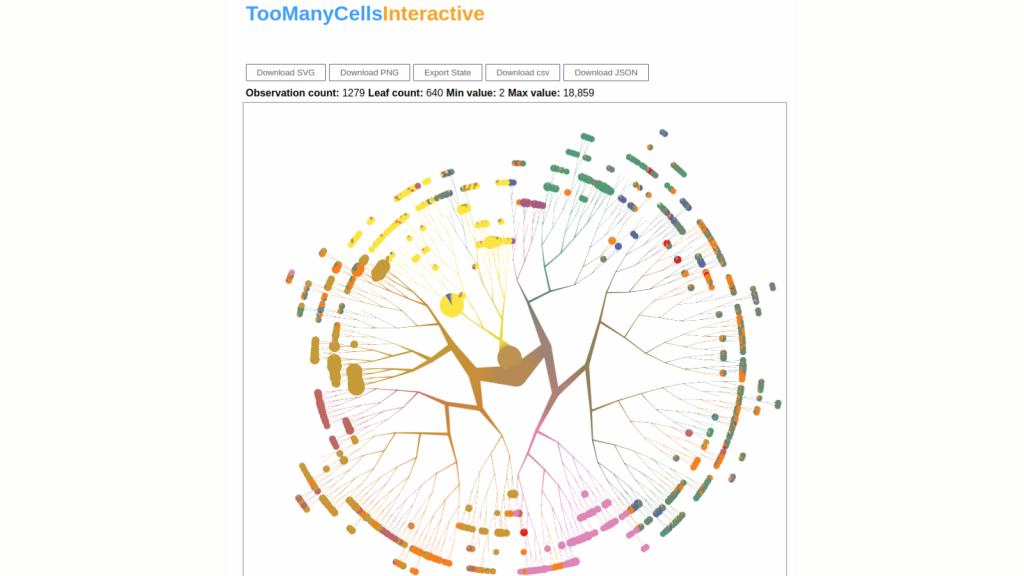
To better understand heterogeneity and drug resistance in cancer, Schwartz and his team developed TooManyCells, a suite of tools designed for clustering and visualizing single-cell data. The visualization component of TooManyCells’ is custom-made and presents cell relationships as a tree. By using TooManyCells, the team could identify rare cancer cells which were contributing to disease progression.
However, the software had some limitations.
“The limitation of TooManyCells was that it took time to build a tree. These trees can be quite large, so to visualize major cell populations you would have to prune the tree several different ways and rerun the program repeatedly. You also didn't really know which way was the right way to prune the tree and colour it until you saw the output,” says Schwartz. “So that’s where this opportunity to work with the DSI’s research software development support program came in.”
“It's wonderful to have a fantastic software developer like Conor devoting his time to facilitating these kinds of projects, which are not easy to get off the ground. They are absolutely necessary and required in these fields but have surprisingly few funding opportunities. So, it's fantastic that these kinds of avenues exist,” says Schwartz about the program.
How is the project developing?
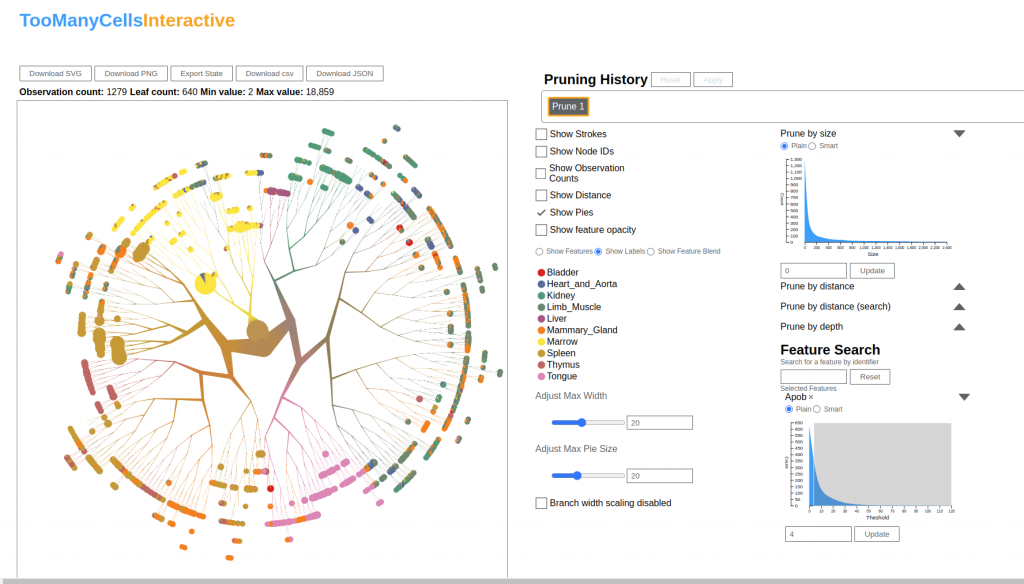
The goal of this project was to provide a graphical user interface for the analysis tools that Schwartz and his team developed. The details have evolved with time but creating an interactive tool to speed up analyses and improve user experience has always been at the heart of the project. Currently, the software development team at the DSI has a prototype in place and is working on collecting user feedback. The research team is also preparing an article describing the software, and once it has been completed, the source code will be made public on the Schwartz Lab GitHub page so that other researchers may access it.
“It's been a pleasure working on TooManyCells! It's given me the opportunity to combine various programming frameworks in ways I haven't done before while supporting some very interesting research,” says Conor Klamann, DSI senior software developer.